Table of Contents
Purpose
The purpose of the METAMOD 2.x software is threefold:
- To manage a local data repository with scientific data files.
- To manage a database with metadata for scientific data sets. Data sets are collections of data files, either in the local repository, or in external repositories.
- To provide a web portal for searching and browsing the metadata database.
As additional projects were seen to benefit from using METAMOD 2.x software, a need for making the software independent of a single project emerged. This was obtained by organizing the software in different modules, and by separating all project specific aspects into a configuration file. METAMOD 2.x now comprise the following modules:
| METAMODBASE | Metadata database and corresponding administrative tools. |
|---|---|
| METAMODSEARCH | Web based search and browsing facility. |
| METAMODUPLOAD | Web based upload facility for data files. |
| METAMODQUEST | Web based questionnaire for registering metadata. |
| METAMODPMH | PMH server offering selected metadata for harvesting by OAI-PMH clients. |
| METAMODHARVEST | OAI-PMH harvester client that import metadata from external sources. |
The configuration file describes an actual implementation of a web application based on METAMOD 2.x software. First of all, the file lists which of the six modules that are used in the application. Any combination of the four modules is allowed. If the METAMODBASE module is not a part of an application, the configuration file must identify a database owned by another application where the METAMODBASE module is used. In this way, several applications may share the same database.
The software is implemented in Perl on a Linux system with Apache web server (as proxy). Catalyst is used to implement dynamic web content, including PMH and other webservices. The database is based on PostgreSQL. Metadata elements use controlled vocabularies from GCMD and CF1.0 as well as WMO keywords. Tools for extracting metadata from NetCDF/CF files are part of METAMODUPLOAD. Access to the actual data files are provided as links from the web pages generated by the METAMODSEARCH module.
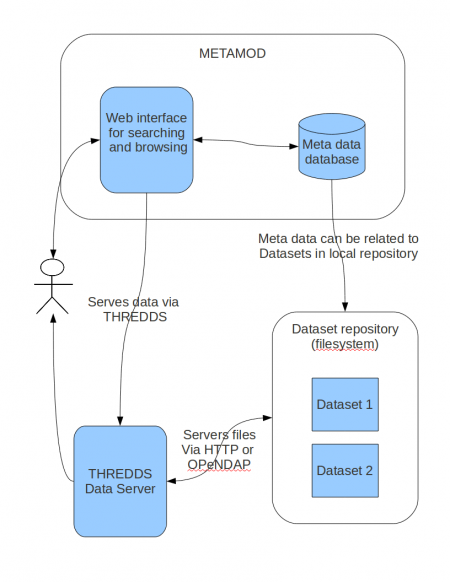
The local data repository is an ordinary file system where files are orginized in catalogs according to institutions (data providers) and data sets. The catalogs and files must be accessible through URLs. This can be done using HTTP, FTP, OPeNDAP or a combination of these protocols. The DAMOCLES application use a THREDDS Data Server (giving OPeNDAP and HTTP access). If the data files satisfy OGC standards, THREDDS may also provide OGC WCS access.
Overview
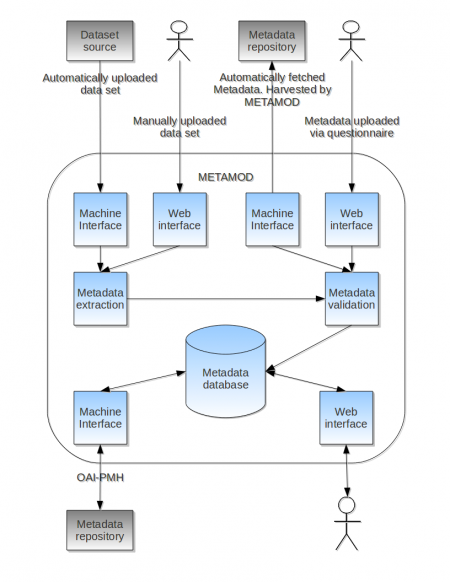
METAMOD is a software system for searching and browsing a repository of metadata for scientific datasets. It consists of a database of metadata, a web interface for searching and browsing metadata and a machine interface that can be used to automatic retrieval of metadata by other metadata repositories.
METAMOD can retrieve metadata from different sources:
- Automatically extracted from netCDF files that are uploaded to the system either manually or automatically.
- Manually added through customizable questionnaires.
- Harvested from other metadata sources via the OAI-PMH standard.
The figure below shows how METAMOD is used to extract, validate and store metadata from different sources. The metadata is then made available to either a human user via the web interface or to other machines via the machine-machine interface. By storing metadata from different sources in the same metadata repository, METAMOD unifies the data available from different sources to simplify finding the desired metadata and associated datasets. An important feature of METAMOD is that it understands several different metadata formats and can convert between the different formats. Currently the following metadata formats are supported: ISO19115/19139, DIF, DC and MM2
In addition to offering a repository of metadata it is possible to extend the functionality of METAMOD by intergrating it with other systems.
- Intergration with the THREDDS Data Server to offer downloading of data sets either via HTTP, OpenDAP, WCS or WMS.
- On the fly interpolation of data before download using the Fimex library
- Web interface for data visualization using OpenLayers (requires WMS server on data-repository)